paired end sequencing wikipedia
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Paired-end tags PET sometimes Paired-End diTags or simply ditags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they theoretically exist together only once in a genome therefore making the sequence of the DNA in between them available upon search if full-genome sequence data is available or upon further.
Now lets get started.
. In addition to producing twice the number of reads for the same time and effort in library. For your De novo genome assembly Fig. The present invention provides for a method of preparing a target nucleic acid fragments to produce a smaller nucleic acid which comprises the two ends of the target nucleic acid.
Therefore they are able to better cover highly. As sequencing projects began to take on longer and more complicated DNA sequences multiple groups began to realize that useful information could be obtained by sequencing both ends of a fragment of DNA. Any platform that can allow for the ligated fragments to be sequenced across the NheI junction Roche 454 or by paired-end or mate-paired reads Illumina GA and HiSeq platforms would be suitable for Hi-C.
Longer runs up to 72nt should be available in early 2009. So you can sequence one end then turn it around and sequence the other end. The paired-end module of the GAII system allows both ends of a DNA fragment to be sequenced.
There are 38 paired end sequencing-related words in total with the top 5 most semantically related being paired-end tags sequence assembly genetics sequencing and dna. Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of. This pipeline is intended for different platforms such as Illumina MiSeq and Illumina HiSeqAlthough this tutorial explains how to apply the pipeline to 16S rRNA amplicons it can be adapted to others markers genespacers eg.
A scaffold is a portion of the genome sequence reconstructed from end-sequenced whole-genome shotgun clones. Briefly the target genomic DNA is isolated and partially digested with restrictio. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
The inner mate distance between the two reads is 200 bp creating an insert size of 400 bp. The reads were then mapped back to the reference using BWA aln and sampe. The unique paired-end sequencing protocol allows the end user to choose the length of the insert 200500 bp and sequence either end of the insert generating highly quality alignable sequence data.
This tutorial describes a standard micca pipeline for the analysis of overlapping paired-end Illumina data. For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter.
Average read lengths for the Roche 454 and Helicos. The paired-end module allows for up to 36 bases of each end to be sequenced during the same sequencing run. A typical paired-end run can achieve 2 75 bp reads and up to 200 million reads.
Run times and outputs approximately double when performing paired-end sequencing. The two sequences you get are paired end reads. What is a scaffold in sequencing.
Read 1 often called the forward read extends from the Read 1 Adapter in the 5 3 direction towards Read 2 along the forward DNA strand. All Illumina next-generation sequencing NGS systems are capable of paired-end. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.
This gives more specificity to alignment algorithms especially in highly repetitive regions. End-sequence profiling ESP sometimes Paired-end mapping PEM is a method based on sequence-tagged connectors developed to facilitate de novo genome sequencing to identify high-resolution copy number and structural aberrations such as inversions and translocations. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
Since paired-end reads are more likely to align to a reference the quality of the entire data set. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. The term paired ends refers to the two ends of the same DNA molecule.
Broader application benefited from pairwise end sequencing known colloquially as double-barrel shotgun sequencing. Before high-throughput sequencing the quality of the library should be verified using Sanger sequencing wherein the long sequencing read. The words at the top of the list are the ones most associated with paired end sequencing and as you go.
The paired-end PE approach where each molecule is sequenced from both the 5 and 3 ends can double the number of bp. This can be very helpful e. Specifically the invention provides cloning and DNA manipulation strategies to isolate the two ends of a large target nucleic acid into a single small DNA construct for rapid cloning.
However in many cases eg with Illumina NextSeq and NovaSeq. Read 2 often called the reverse read extends from the Read 2 Adapter in the 5 3 direction. Output per run for single-end sequencing are noted.
High-throughput sequencing of 16S rRNA gene amplicons is a valuable tool for comparing microbial community structure among hundreds of samples for which researchers have primarily used the 454 Pyrosequencing platform. You can get the definitions of a word in the list below by tapping the question-mark icon next to it. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1.
Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost-effective manner and that the additional benefits obtained from paired-end sequencing are not worth the additional cost. Since paired-end reads are more likely to align to a reference the quality of the entire data set improves. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
The larger inserts mate pairs can pair reads across greater distances.

Whole Genome Sequencing Si Pengawas Mutasi Virus Genecraft Labs
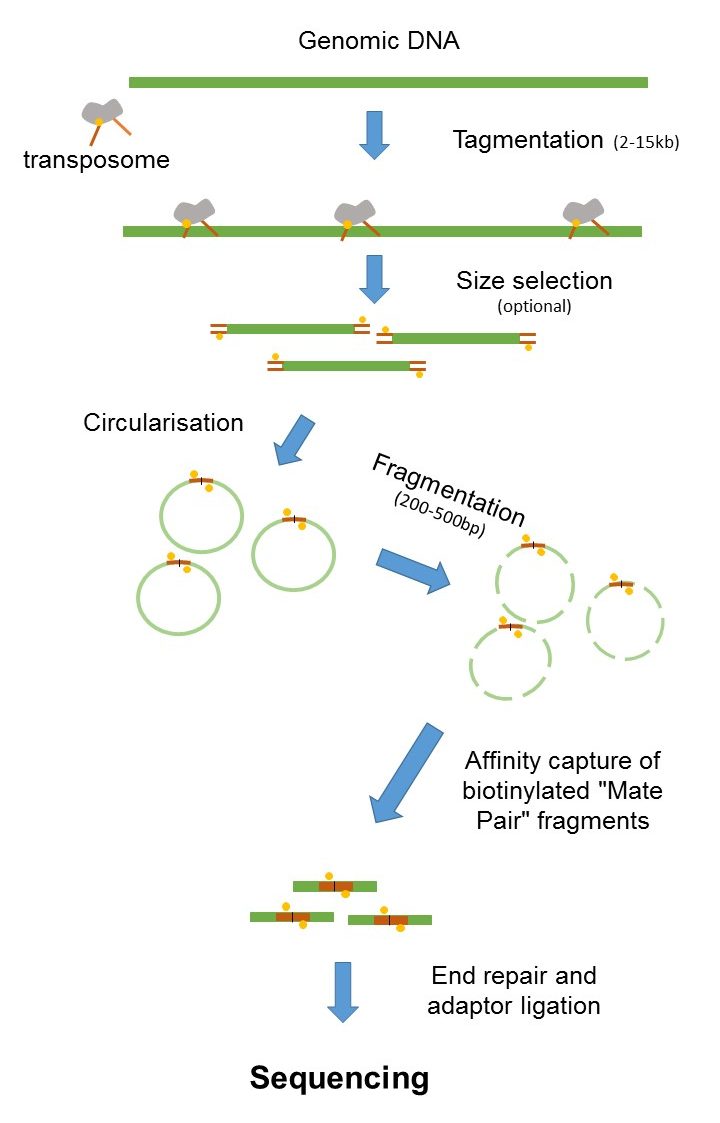
Mate Pair Sequencing France Genomique

Bacterial Chromosome Replication Biology Lessons Chromosome Biology
Trehalose Increases Tomato Drought Tolerance Induces Defenses And Increases Resistance To Bacterial Wilt Disease
Informatics For Rna Sequencing A Web Resource For Analysis On The Cloud Plos Computational Biology

Hi C Genomic Analysis Technique Wikipedia

Hybrid Genome Assembly Wikiwand

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist

Bacterial Chromosome Replication Biology Lessons Chromosome Biology
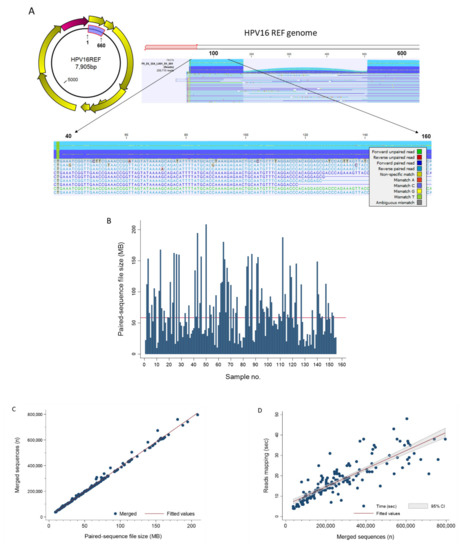
Pathogens Free Full Text Hpv Deepseq An Ultra Fast Method Of Ngs Data Analysis And Visualization Using Automated Workflows And A Customized Papillomavirus Database In Clc Genomics Workbench Html

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For






